timsTOF HT
High-throughput 4D-Proteomics™ High dynamic range and quantitation accuracy

timsTOF HT
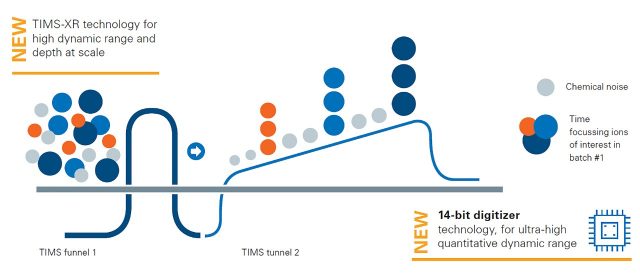
Powered by the 4th generation TIMS-XR and advanced digitizer technology for high dynamic range and analytical depth in quantitative cell and tissue proteomics.
Expanding the capabilities of high-throughput 4D-Proteomics™
TIMS based ion mobility separation and CCS-enabled analysis
The new timsTOF HT mass spectrometer is equipped with the 4th generation high capacity TIMS-XR analyzer and advanced digitizer technology (ADT). It offers higher dynamic range for unmatched analytical depth and quantitation in high-throughput proteomics experiments. The timsTOF HT is fully CCS-enabled and supports all PASEF® acquisition modes including PASEF®, dia-PASEF® and prm-PASEF® achieving flexibility in 4D-Proteomics™. Simply uncompromised proteomic depth at scale.
Bruker ProteoScape™
Maximize your identification depth and throughput with the new capabilities of Bruker ProteoScape™, our “run and done” database search platform. TIMScore™ and TIMS DIA-NN take full advantage of the CCS for more accurate protein identification on the fly.

Enabling rapid tissue proteome analysis with dia-PASEF®
Rapid cardiac tissue proteomics. Different amounts of a murine heart proteome digest were analyzed in triplicates with short liquid chromatography gradients and dia-PASEF®. At the highest sample load, we identified in total 4700 protein groups spanning more than five orders of magnitude in relative abundance. Raw data were processed with DIA-NN in library-free mode and protein copy numbers were estimated with the ‘proteomic ruler’ approach.
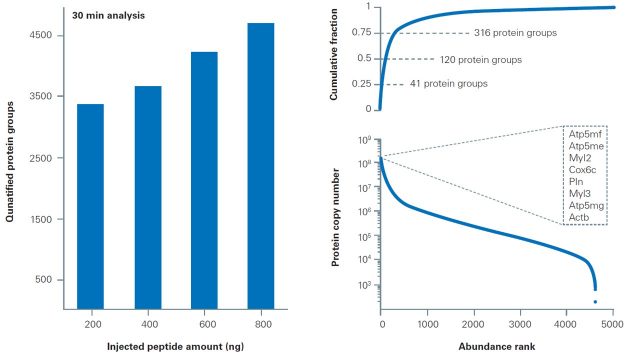
"Tissue analysis is a key area in clinical proteomics, yet extremely challenging as tissue sections and biopsies comprise a vastly heterogenous cell population. The dia-PASEF® acquisition mode on the timsTOF HT instrument quantifies proteins across a large dynamic range even in notoriously difficult samples such as cardiac tissue without sacrificing throughput or sensitivity."

Unprecedented depth in cell lines with dia-PASEF®, TIMS DIA-NN and Bruker ProteoScape™
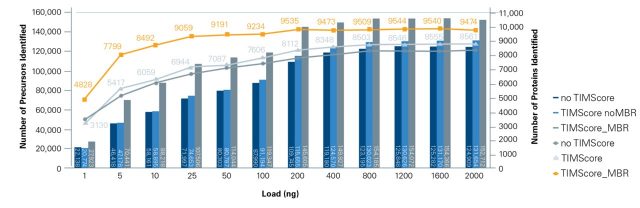
For even higher proteome depth, increased protein digest loads improve identifications. A tryptic digest of human cell line K562 (Promega) was analyzed using a nanoElute® system at 250 nL/min with 60 min separation time on an Aurora column (25cm, 75 μm ID, IonOpticks). timsTOF HT operated in dia-PASEF® mode using 100 ms TIMS separations. Data was processed with TIMS DIA-NN using a predicted library from uniprot (human reviewed).
Targeted Plasma Proteomics: quantify more than 550 proteins with a 30 min gradient
We combined the use of a PQ500 (Biognosys) Stable Isotopic Standard (SIS) with a dia-PASEF® based appoach using a 30 min gradient on a timsTOF HT. 578 proteins (corresponding to 804 peptides) could be accurately quantified with a > 99% data completeness. This approach combines the selectivity and sensitivity of traditional SMS/MRM methods with an easy-to use acquisition setup, also allowing to define new targets a posteriori.
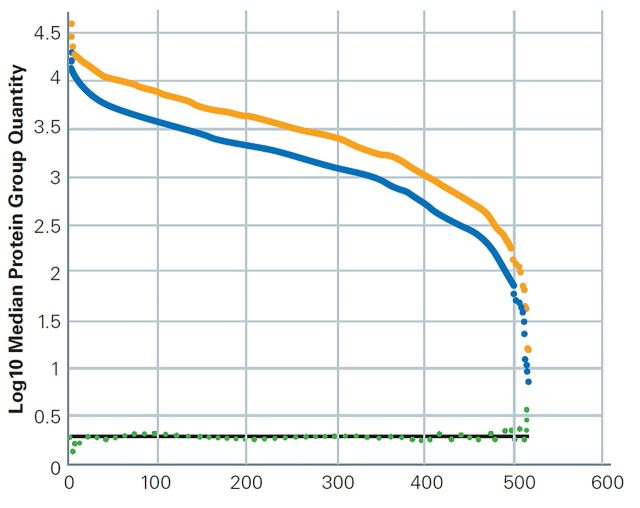
Nano LC columns for best results on higher sample loads
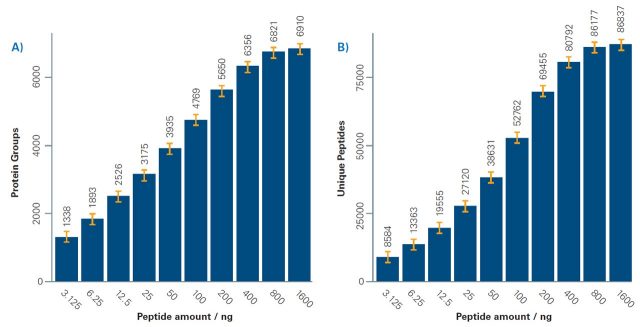
A) The TIMS-XR allows for increased ion capacity for sample loads beyond 200 ng. Reproducible identification of ~ 6900 protein groups and more than 86,000 unique peptides.
B) Number of identifications using the Bruker ProteoScape™ realtime search engine with TIMScore™ for dda-PASEF of up to 1600 ng K562 tryptic digest. Error bars indicate standard deviation for 5 replicates with mean displayed above histogram.
Using a PepSep™ Twenty-five series column (150 μm ID, 1.5 μm particle size), average peak of widths less than five seconds were achieved for K562 tryptic digest (Promega) sample loads up to 1600 ng. A gradient time of 41 min was adequate for efficient peptide separation, resulting in homogenous peptide elution across the gradient even at high peptide loads.
"Our ongoing collaboration with Bruker to tailor DIA-NN to a streamlined processing tool for dia-PASEF® data with a CCS focus has been rewarding. It simplifies and accelerates identifying and quantifying thousands of proteins in very short gradients. We are pleased that within our close collaboration with Bruker, the vendor-integrated version of DIA-NN called TIMS DIA-NN now becomes part of the Bruker ProteoScape™ bioinformatics platform."













