impact II VIP
Analytical performance for trace analysis in complex matrices
Ultimate sensitivity
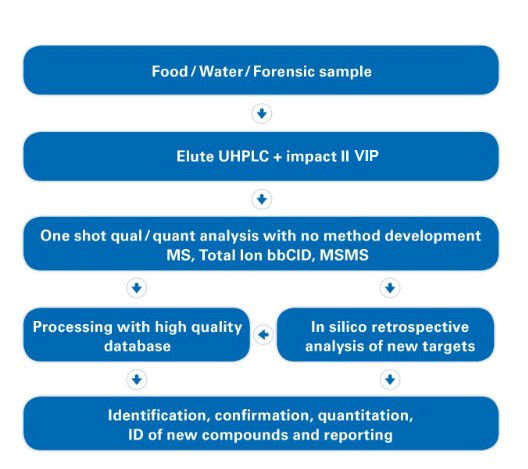
One run for qual and quant

impact II VIP
Screen residues and contaminants, identify impurities
Enabling chromatography
Even outstanding mass spectrometers benefit from good chromatography. By choosing the impact II VIP, hyphenation is made easy with
- UHPLC
- GC
- CE
- IC
Switching from one modality to another takes less than 10 minutes each.
Fast methods offer additional flexibility and do not necessarily require a chromatographic device. The Direct Insertion Probe serves best.
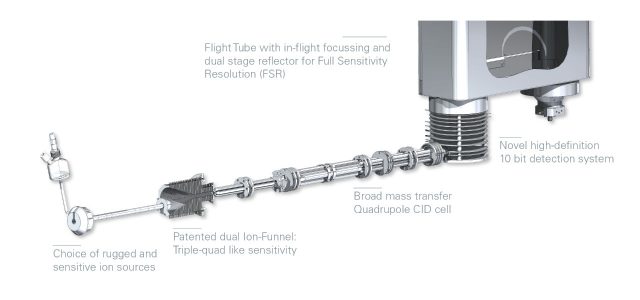
Instant expertise in accurate mass LC-MS/MS
Research-grade technology applied to every routine analytical task. The Bruker impact II VIP QTOF features:
- Low picogram sensitivity while delivering, highly precise MS and MS/MS data
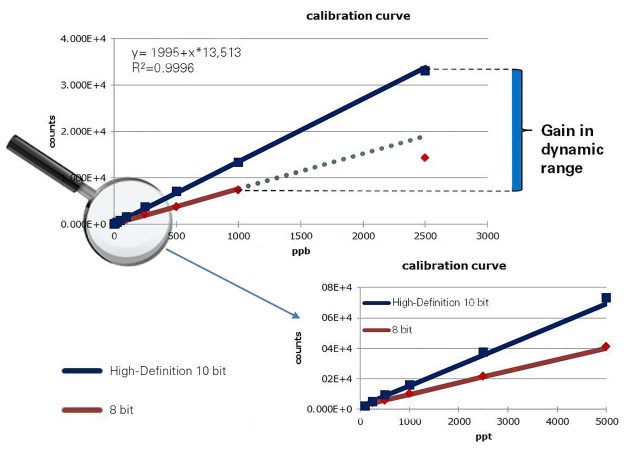
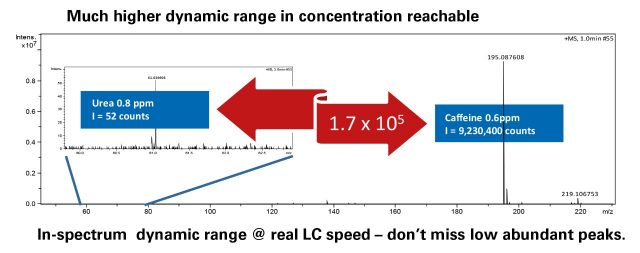
- Definitive trace analysis from complex, high-background matrices, by increased dynamic range
- True Isotopic Pattern for unambiguous spectra
- Superior 1 ppm mass accuracy ensured by automated calibration
- 50 GBit/sec sampling technology maintaining full specification resolution even during ultrafast chromatography
The impact II VIP combines the robustness of Bruker orthogonal Time-of-Flight instruments with 10-bit digitizer technology that allows for compound detection and identification across an unrivalled dynamic range.
Achieve your goals in a range of applications
- Environmental and pollutants analysis
- Food, feed and beverage QC
- Forensics and doping control
- Drug metabolite, degradant and impurity identification and quantitation
- Biomarker discovery and validation in proteomics and metabolomics
- Intact protein analysis and characterization of biopharmaceuticals
High-performance OMICS
High performance proteomics and metabolomics studies using the impact II VIP QTOF provide insight into how rational strain design increased bio-production efficiency. The Bruker impact II VIP QTOF system is well suited to bottom-up proteomic and metabolomic workflows, and can be used to map the results of both metabolic and proteomic changes onto biochemical pathways.